LatchBio
Fifty Years partners with LatchBio to put the power of bioinformatics in the hands of every biologist.
Biotech is drinking through a firehose. Next-generation sequencing, CRISPR, and the high-throughput revolution have generated massive datasets that hold the secrets to hacking life. These methods are becoming easier and more standardized every day. While the velocity and volume of biological data is exploding, our ability to analyze it all is not.
Chewing through these databases is no easy feat. Biological data is inherently messy because biology itself is messy. Our high school textbooks led us to believe that life operates on a set of deterministic processes that can be predicted – coding DNA will be transcribed into RNA, plants will turn CO2 into biomass, and specific chemicals will give you cancer.
But in reality, biology is sampling from a distribution of potential outcomes that can be analyzed statistically but may not be precisely predicted. At the cellular level, there is only a chance DNA is transcribed, CO2 is fixed, or a cell is induced to become cancerous. This lack of precise outcomes – or stochasticity – underpins any biological experiment and therefore the resulting data.
The difficulty of finding signal in the noise has only increased with modern technology. Until very recently, biologists would measure single readouts of their experiment. For example, they would hit cancer cells in a petri dish with a drug and measure how a single gene changed compared to unperturbed cells. But now, we use unbiased approaches that measure all gene expression changes using sequencing. Controlling for experimental error and stochasticity to generate inferences was hard enough when measuring one gene – it has become exponentially harder with measuring thousands.
The messiness and size of these datasets created a need for bioinformatics – a specific skill set that is required to turn -omics data into biological knowledge. Bioinformaticians spend years understanding the rigorous statistics that allow inferences to be made. They master the R libraries and Python packages that have been developed to comb through data. And they form the computational chops that turn one-off scripts into full bioinformatic pipelines or a folder full of sequencing data into an all-out data warehouse.
But herein lies a chicken or egg problem, the catch-22: obtaining the -omics data also requires a ton of training completely orthogonal to bioinformatics. Biologists hone their skills working with the stochasticity generator that is life to create well-designed experiments that feed these vital databases.
But most biologists don’t have the tools to do bioinformatics. And most bioinformaticians have never held a pipette. Both of these non-overlapping skill sets are essential to both leverage the newest biotechnologies and sift through the data created.
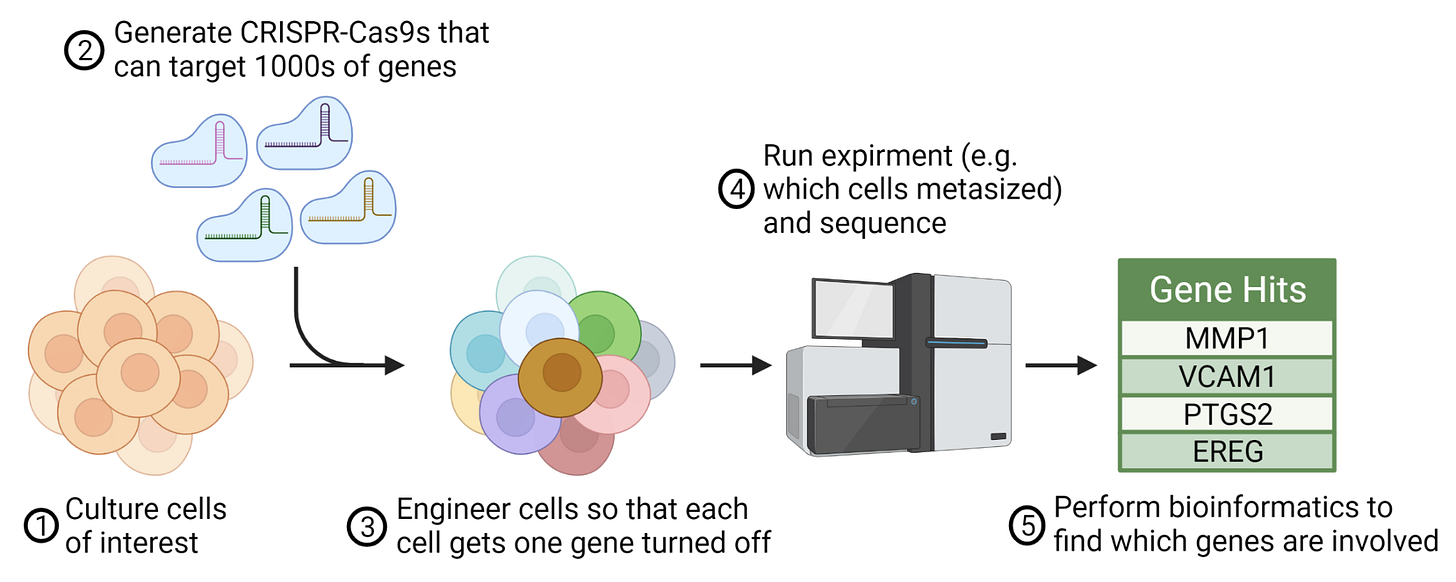
Let’s take an example using the hottest tool in biology – CRISPR. These molecular scissors allow us to make targeted edits anywhere on the genome, effectively turning off any gene we desire. The work that has generated a Nobel prize and multiple breakthrough therapies has birthed CRISPR screens; an entirely new method that combines sequencing, CRISPR, and high-throughput biology. Here, a pool of cells are edited in thousands of different locations on the genome, one edit in one cell, turning off each gene. Combining experimental and computational approaches, scientists can ask questions like “which genes are essential for cancer to metastasize?” or “which genes help T cells inhibit autoimmunity?”.
To pull this off, biologists leverage their skill sets to learn how to grow the cells of interest, get CRISPR in the cell, and create an experiment that tests the question – all before even starting the screen! While holding most of the parameters constant, they use CRISPR to precisely disrupt a single gene at a time and carefully measure if this perturbation had a biological effect. The process of doing this for thousands of genes is called a genome-wide "screen" and allows a rich characterization of behavior over the whole genomic landscape. Once the experiment is complete, they rip out the cell's genetics and submit it for sequencing.
With data hot-off the sequencer, the bioinformaticians then take over. Sequencers produce files that are too large to look at and aren’t very human readable. They contain hundreds of millions of small stretches of genetic information that were pulled from the cells and now need to be identified and quantified.
Bioinformaticians use algorithms to map these reads back to places on the human genome, run data pipelines that connect these reads to specific cells, and execute code that tells them which cell got which edit and how much of each gene was expressed in a single cell. Each step requires a new pipeline and new understanding of the statistical framework that makes it work – all to produce a matrix of cells by gene that can be further analyzed and experimentally validated.
The training needed to pull off either the wet or dry portion of a single CRISPR screen is enormous. There are very few people who are excellent at both, creating a problem for anyone looking to leverage this method.
So, should a company, lab, or institute use limited resources to hire biologists to generate data without the ability to effectively analyze it? Or hire bioinformaticians and sacrifice on the ability to generate the data goldmine that will drive the agenda forward?
We think they shouldn’t have to choose. Enter LatchBio.
As undergraduates at UC Berkeley, LatchBio co-founders Alfredo Andere, Kenny Workman, and Kyle Giffin recognized how many potential life-saving therapies, climate improving biomolecules, and novel ways to hack biology were sitting undiscovered in datasets that humanity didn't have the bandwidth to dive into. Taking the deep expertise in computer science they had gained from working at companies like Asimov, Facebook, Google, and Serotiny; leading Machine Learning at Berkeley; and their bench experience at places like the Joint BioEnergy Institute and the Lawrence Livermore National Laboratory, the trio left their degrees behind and founded LatchBio.
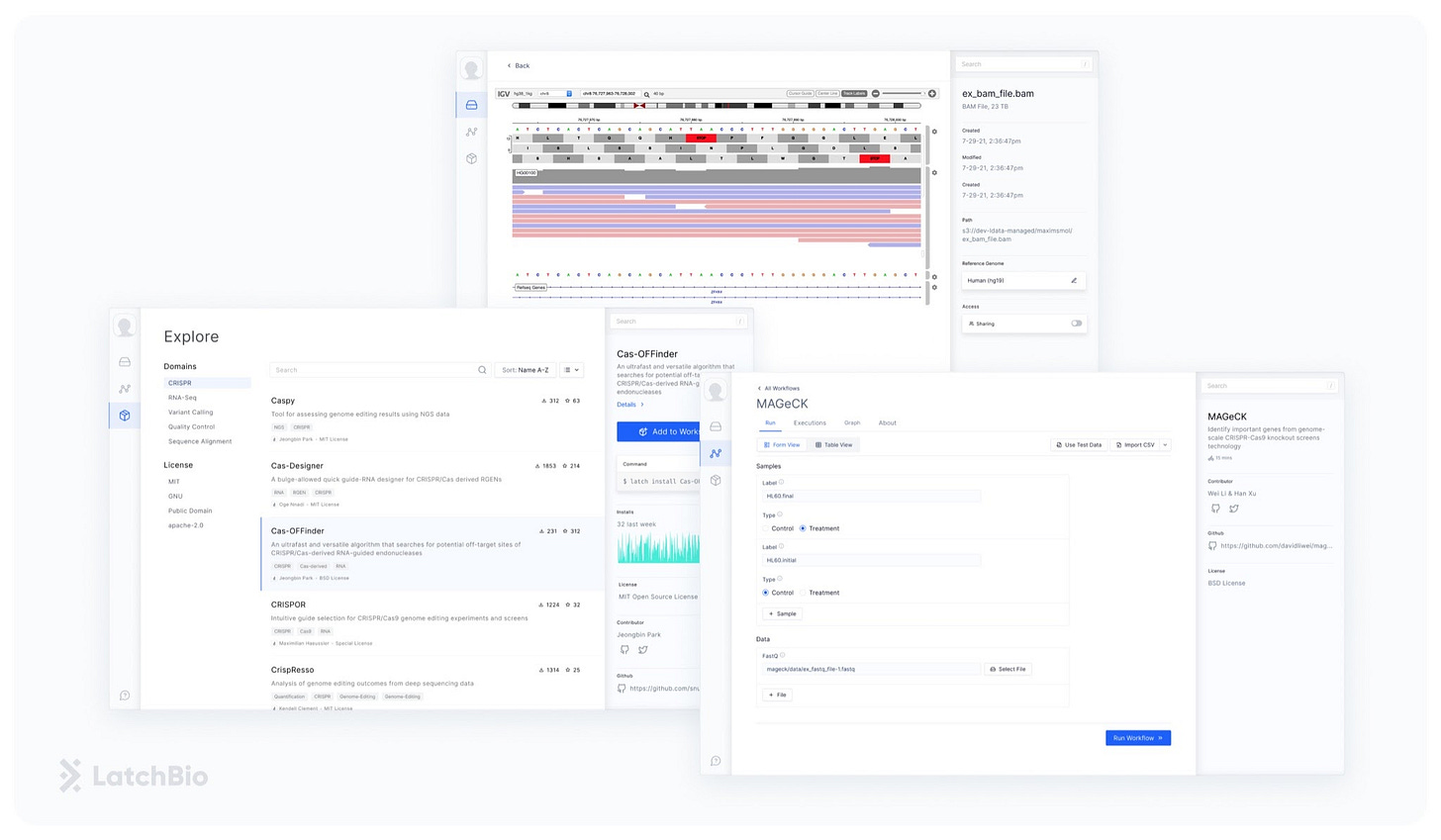
LatchBio’s platform puts analysis back in the hands of biologists, enabling them to leverage the explosion of data they’re generating. LatchBio created intuitive out-of-the-box tools that abstract away all the arcane statistics, code, and computational backend that inhibit biologists from working with data, allowing anyone to build their own data warehouses, analytical tools, and bioinformatics pipelines.
In the CRISPR screen example, any biologist using LatchBio’s platform can upload their data directly from the sequencer with a click of a button. Sequence QC, sequence-to-genome alignment, gene expression quantification, and edit site identification are all run in the cloud without the biologist even knowing – all they see are simple-to-use drag and drop tools neatly assembled into a flow diagram. Data storage and visualization are also just as easy to do, all taken out of a terminal and into a user-friendly web interface.
But don’t worry bioinformaticians – LatchBio is not looking to make you obsolete! Instead, LatchBio wants to empower bioinformaticians to focus on making the tools that give biological data meaning. Instead of wasting time running the same code over and over for biologists, bioinformaticians will be able to use LatchBio’s platform as the next generation of GitHub for computational biology. Unlike current software repositories, the workflows generated through LatchBio will be immediately accessible to biologists, ensuring tools don’t have huge learning curves until they become widely adopted. This accelerates software development by creating a space where biologists with data and bioinformaticians with tools under construction can collaborate and support each other.
And they won’t stop there. LatchBio is initially focused on doing the types of CRISPR workflows described above to accelerate the reach of these important therapies. In the future, they’ll incorporate every workflow computational biology has developed, from single-cell sequencing to metabolomics to phylogenetics, all under one roof. They’ll generate out-of-the-box machine learning to democratize access to these even more challenging algorithms. And they’ll streamline data sharing so well that with a few clicks, data can be reproduced from published work and publicly available databases.
In a future where LatchBio succeeds, all compute power the world has to offer will be leveraged to analyze biological data. LatchBio’s web-based platform will make it incredibly easy and seamless for bioinformaticians to develop and biologists to deploy bioinformatic workflows. By tearing down silos of knowledge that currently inhibit progress, LatchBio can leverage every idle CPU to uncover hidden connections in disparate data centers, radically accelerating biotechnology and medicine.
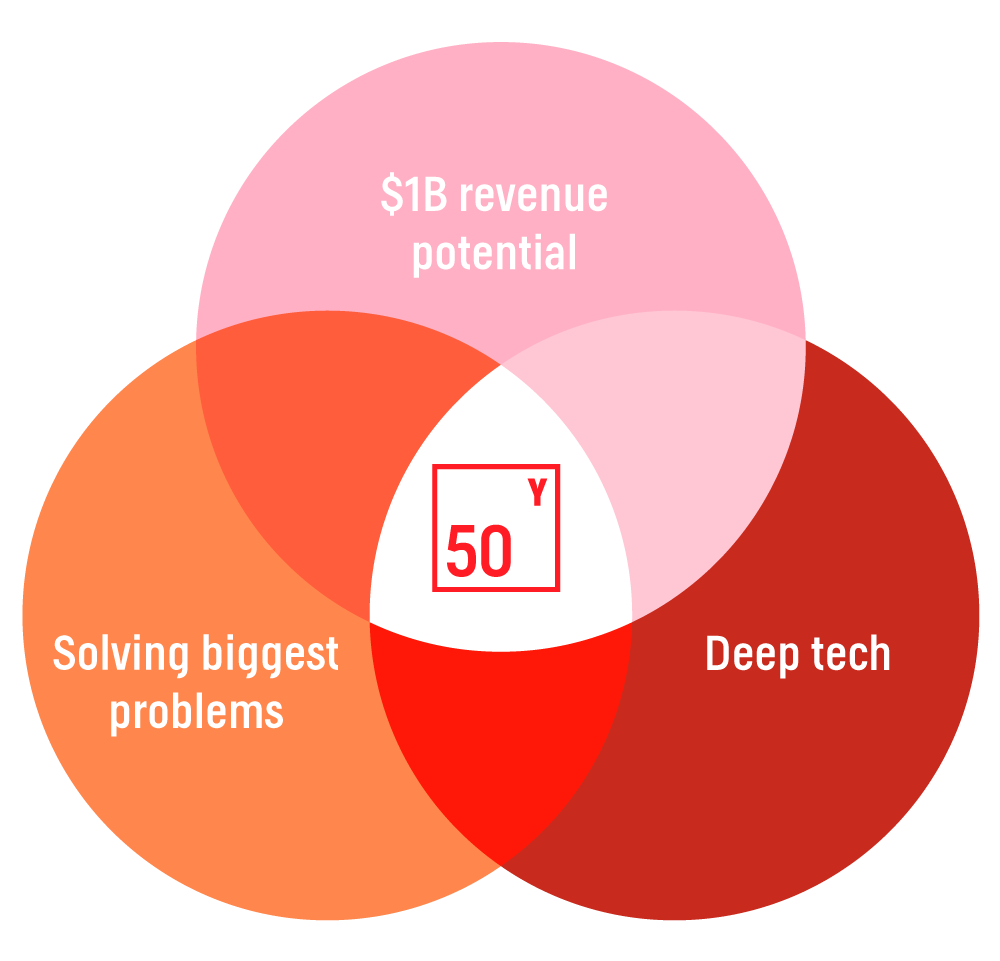
At Fifty Years, our sweet spot is supporting founders at the earliest stages building deep tech companies that can generate huge financial outcomes and create massive positive impact.
Deep tech: Alfredo, Kenny, and Kyle are leveraging their technical expertise in computer science, computational biology, bench science, and machine learning to create a platform that is accessible to everyone in the biology stack.
$1B yearly revenue potential: Every single new biology endeavor must hire a computational biology team to build out data-warehouses, analytical tools, and bioinformatic pipelines just to make use of their data, effectively reinventing the wheel each time. LatchBio fills this need, serving anyone who uses biological data – virtually every biology lab, institute, and company.
Massive positive societal impact: Currently, data could take weeks to analyze as access to bioinformaticians and compute resources hinder the process. LatchBio democratizes access to these tools, turns this into minutes, and accelerates the development of more sophisticated tools. This means life-saving therapies and environment healing tools are identified exponentially quicker.
Inspired by their technology and vision for the future of computational biology, Fifty Years is excited to partner with LatchBio in their seed round led by our friends at Lux Capital, with participation from General Catalyst, Haystack, Brian Naughton, and computational biology founders. We’re looking forward to helping Alfredo, Kenny, and Kyle lay the bricks for the biocomputing revolution.